– by Mustafa Chasmai and Rohan Garg
Introduction to Protein Folding
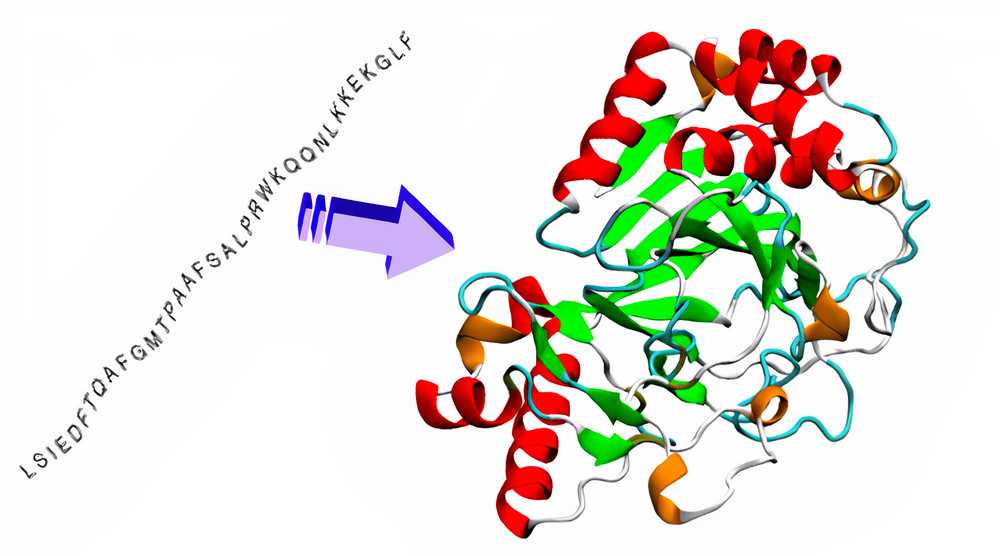
Proteins are integral building blocks of life. Their complex functions and rich set of reactions largely depend on their structure. When relatively simpler poly-peptide chains are ‘folded’ in specific configurations, specific biologically active proteins are formed. But it is very difficult to tell which configurations would lead to which property, and so, to tell the configuration from the properties. The latter is the crux of the 50 year old problem of protein folding.
The difficulty of this problem, as amply demonstrated by Levinthal’s Paradox, lies in the fact that there are an estimate of 10300 different configurations a single protein can have. Over the past few decades researchers have been able to determine shapes of various proteins using different methods, but each method depends on a lot of trial and error, which can take years of work, and cost tens or hundreds of thousands of dollars per protein structure. Interestingly, these techniques don’t even work for some types of proteins.
Google DeepMind
But why are we rambling on about biology in this ML-related blog? Using ML, scientists at DeepMind were able to finally solve this half-a-century old problem. Their model ‘AlphaFold’ won the Critical Assessment of protein Structure Prediction (CASP) global competition, which is considered to be the gold standard for assessing protein predictive techniques. AlphaFold may help rare disease researchers predict the shape of a protein of interest rapidly, eventually contributing to efficient and economical drug discovery.
Their Approach
Their method relies on deep neural networks that are trained to predict properties of the protein from its genetic sequence. They used a combination of unlabelled and the relatively small labelled data. Using the much larger unlabelled data, they extracted embeddings (vector representations) and used embeddings of labelled data for training the primary networks. The properties their networks predicted were: (a) the distances between pairs of amino acids and (b) the angles between chemical bonds that connect those amino acids. Using this, they are able to deduce the complex 3D structure of proteins with a very high accuracy, many of them being correct within 1 atom’s width.
DeepMind is known to have trained the program on over 170,000 proteins from a public repository of protein sequences and structures. They used a form of attention networks, where they effectively broke the problem into smaller parts, solved them separately, and then finally brought it all together. A detailed understanding of their approach will be possible only after the official paper gets published.
Other Applications of AI / ML in Fundamental Sciences
Besides protein folding, AI/ML is finding applications in many other fields that at first glance, are not remotely related to ML. One such awesome application is in Basic Maths. Imagine being able to prove Zorn’s lemma instantly. Imagine an ML-based algorithm finally solving the 160-year old Reimann hypothesis.
Protein folding is just one of the many many applications of AI/ML. From microscopic proteins in our genes to massive black holes in the universe, ML is affecting people's lives in many different fields. Exploring these applications may lead us a step closer to unravelling the secrets of general AI.
In the second episode of our ML Muse podcast series, we had a talk with Ankit Anand from DeepMind, Montreal. He has a PhD in AI from IITD and is currently working with DeepMind on many problems including some applications in Basic Maths. Through questions on his journey and work, we show a possible path each and every one of us could take. We discuss his journey, his thoughts about the research culture at IITD and at DeepMind and some general advice for us young students starting out in this interesting and sometimes truly humbling field.
To know more about the podcast, visit the post or watch it on YouTube!

